Antony N. Daviesa and Luc Patinyb
aSERC, Sustainable Environment Research Centre, Faculty of Computing, Engineering and Science, University of South Wales, UK
bZakodium Sàrl, route d’Echandens 6b, 1027 Lonay, Switzerland
DOI: https://doi.org/10.1255/sew.2021.a18
© 2021 The Author
Published under a Creative Commons BY licence
As you will have seen in recent columns there has been much discussion about what spectroscopic data format is most appropriate in different scenarios. Depending on your workflows. an image might be the best format even if it hurts the “standardisation at all costs” side of my brain to admit it (for example see Figure 2 in Reference 1). During part of the quite animated discussions on this topic. especially around making repositories future-safe by ensuring data processing, Damien Jeannerat asked me if I had seen the release of a free nuclear magnetic resonance (NMR) web-based data processing tool which could handle both raw data files as well as JCAMP-DX NMR standardised files. This tool is NMRium, produced by Luc Patiny and colleagues at a Swiss scientific data management company called Zakodium. I had not heard of it and so I looked at their webpage and was impressed, so asked Luc directly if he was happy to be the subject of a column in SE to discuss the interesting innovative solution they are now providing users for free. (Just a quick reminder that the “F” in FAIR does not standard for Free or even open access!)
Luc was very forthcoming about the NMRium system from its beginnings and the ethos behind the open source development work that their company had done. I hope you enjoy his comments and try out their system. I am certain they will welcome any feedback you may have or suggestions for functionality!
Background to the NMRium project—why would a company want to produce an advanced data processing application and make it available for free?
The owners of Zakodium are all scientists and we maintain and participate in over 150 open source projects (several of them can be found in areas such as machine learning, image and data processing at https://www.zakodium.com/open-source). An early driver was that, as scientists, we found out that far too much data are lost because they are not stored and shared correctly. When carrying out research for our 2016 paper2 we found out that it was difficult to find open access NMR spectra. However, the results showed that we could clearly produce a self-learning algorithm that would be extremely powerful IF more data were available.
One of the reasons why data are not available is that software to reprocess spectra are expensive and (as published in this column in recent editions—Ed!) some people only make available PDFs or static images of their spectra. There is also no straightforward workflow that allows the storage and sharing of data and share it (and Damien’s NMReData format is one solution).3
We were also looking to NMRshiftDB, and in their database there are nearly no JCAMP-DX files, only the chemical shifts.
A grant from the German government4 enabled the NMRshiftDB team to develop a new way to process NMR data in the browser. We took the lead to develop such a solution as an open source React component.
The overall NMRium project is a collaboration developed between Zakodium Sàrl, Switzerland, the University of Cologne, Germany, Johannes Gutenberg University Mainz, Germany, and Universidad del Valle, Colombia. The project was funded by the IDNMR DFG grant, as well as Zakodium Sàrl and the Universidad del Valle, Cali, Colombia.
Zakodium Sàrl is I assume a commercial company—is it the intention to start getting people to pay for this service?
Zakodium is a company that specialises in the storage and processing of scientific data with the goal to convert data to knowledge. The service of processing NMR spectra will stay free (anyway it is an open source project, you could just take a copy of the component). Our paid services are consulting, custom development and data management.
When you say that there is no backend processing on a server, so it is “Safe” is there any data stored on the web server or is all processing carried out in local memory—so effectively only on the local PC?
We took care that ALL the processing is done in your PC and nothing is sent to the server. As an experimental feature you can even install it locally on your PC (as a PWA, progressive web app) and you will be able to process NMR spectra offline. There is a small icon on the website that allows to do this (Figure 1).
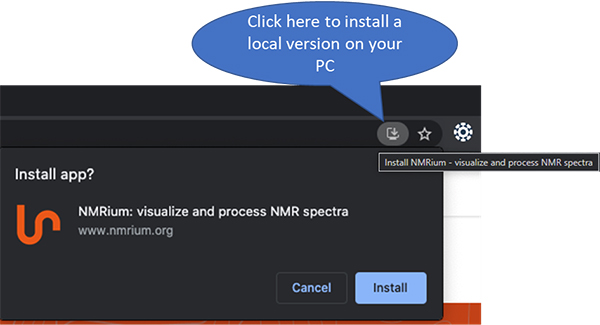
Figure 1. This is a screenshot taken using an up-to-date Chrome browser under Windows 10.
Even if you access the spectra using this kind of URL, https://www.nmrium.org/nmrium#?jcamp=https://cheminfo.github.io/nmr-dat…, we take care to add this “#” so that your browser does not send the URL of the JCAMP-DX file to our server. (Ed.: I had to look this up! In JavaScript, the query after the # character is only handled by the client side of the service running locally on your machine and not parsed back to the server.)
How would you describe the state-of-the-art environment which you have now improved on by the release of NMRium?
We are convinced that in the future the only application you will need on your computer is a web browser. In fact, your computer, after 40 years, is again becoming a “terminal” (you process data that are not locally saved from a browser). While today it is easy to have spreadsheets, word processors or email in the browser, the processing of spectra is barely possible. This means that up to now you had to install an application on all the computers that are required to process spectra. Installing software is expensive not only for licenses but also the cost of IT to install the software (in some companies it is really complex).
By using web applications, you avoid all the problems of installation and updates (simply reload the page and you get the latest version).
NMRium in action
OK thanks Luc, so let us see NMRium in action. When you open the program, you are faced with a large area of whiteness with some clear instructions on how to proceed (Figure 2).
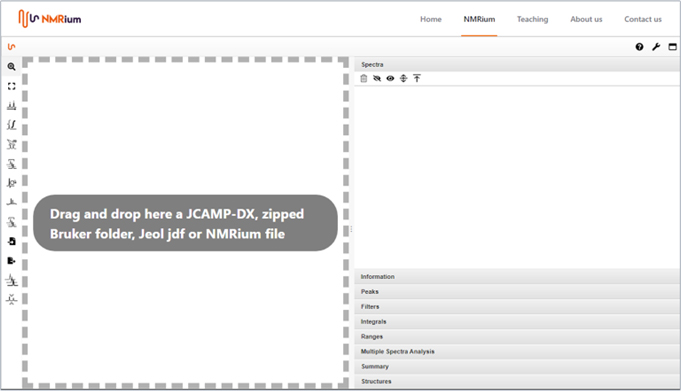
Figure 2.
For those of us who do not read instructions and just try to dump the NMR files into the program they nicely remind you with some more details of what you really need to do (Figure 3).
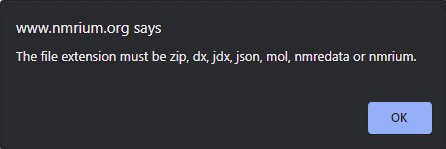
Figure 3.
Suitably embarrassed by my overconfidence (who does ever read QuickStart guides by the way?), I decided to test the system out with Peter Lampen’s original JCAMP-DX encoding test files, which are 1D ethylbenzene NMR spectra and an FID encoded using the various allowed JCAMP-DX XY-DATA encoding standard algorithms AFFN, PAC, SQZ, DIFF and NTUPLES formats. As you can see from Figure 4, NMRium passed with flying colours showing that NMR data which was originally saved in 1992 using the JCAMP-DX NMR 5.01 standard format can still be read almost 30 years later.
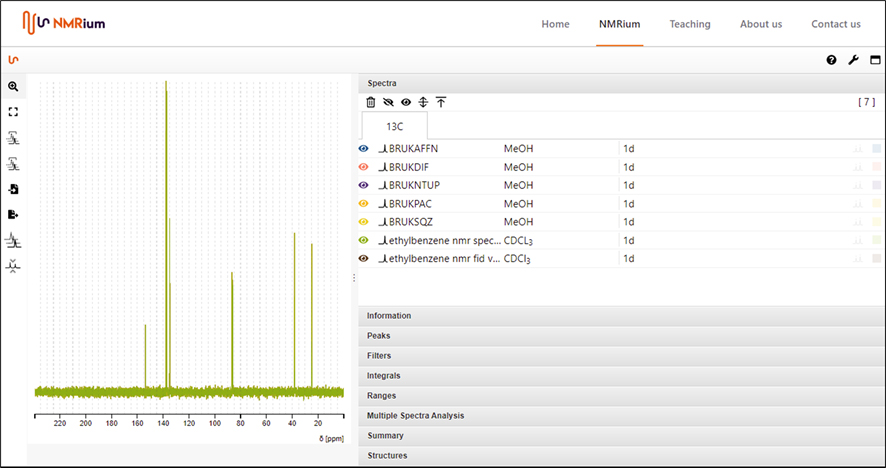
Figure 4.
There are all the usual NMR data processing tools you would expect including 1H-NMR prediction which I carried out for Robert Lancashire’s favourite acetophenone but I have run out of space for more figures.
Conclusions
I am very pleased that new Open applications are becoming available against which we can test the FAIR principles and data longevity strategies that have been developed over the years.
Tools like NMRium will greatly help in moving to a FAIR world as it means organisations that cannot afford to pay acquisition and maintenance contracts on expensive specialist spectroscopic software will still be able to get quite detailed access and analysis of spectra stored in Open spectroscopic data repositories.
Damien’s vision for this capability goes further, “anybody” can make a set of spectra available on a mini web server, a GitHub page in fact. He published an example in his most recent paper with Carlos Corba from Mestrelab Research.5 It is easy to imagine having a mini site for Zenodo, Dataverse etc. repository including NMR. If these repositories then had an agreed, open and uniform structure, this could be fully automated (eliminating some of the manual steps we had in the EuroSpec spectroscopic repository).
Creating such a dataset involved dropping a spectrum into NMRium, then using a Save As… cycle to create the JCAMP-DX file (something that could be easily automated using an archive forge of a simple front-end page—yeah, I keep talking about this!). Then a script automatically generates a table of content file that ends up as the anchor of the URL. Pretty elegant stuff!
The NMRium format allows spectra to include embedded links. Nice to avoid duplicating data. Ideally, if the link could extract an individual file from a zip file sitting in a repository, it would be a dream come true for NMR archaeology!
In fact if you want to see more there is a short explanatory video below and at https://www.nmrium.org/videos/presentation. Better examples than I have created for the column including 2-DNMR are available at https://www.nmrium.org/nmrium#?toc=https://cheminfo.github.io/nmr-dataset-demo/samples.json. You can also download the demo data from https://github.com/cheminfo/nmr-dataset-demo to try out the entire process. By the way note that the example URL launches NMRium but loads the data set it is showing from GitHub. Damien, your dream is close to being reality!
Everyone please, stay safe!
References
- R.M. Hanson, D. Jeannerat, M. Archibald, I. Bruno, S. Chalk, A.N. Davies, R.J. Lancashire, J. Lang and H.S. Rzepa, “FAIR enough?”, Spectrosc. Europe 33(2), 25–31 (2021). https://doi.org/10.1255/sew.2021.a9
- A.M. Castillo, A. Bernal, R. Dieden, L. Patiny and J. Wist, “Ask Ernö: a self-learning tool for assignment and prediction of nuclear magnetic resonance spectra”, J. Cheminform. 8, 26 (2016). https://doi.org/10.1186/s13321-016-0134-6
- S. Kuhn, L.H.E. Wieske, P. Trevorrow, D. Schober, N.E. Schlörer, J.-M. Nuzillard, P. Kessler, J. Junker, A. Herráez, C. Farès, M. Erdélyi and D. Jeannerat, “NMReData: Tools and applications”, Magn. Reason. Chem. online ahead of print (2021). https://doi.org/10.1002/mrc.5146
- IDNMR Grant. Part of the Scientific Library Services and Information Systems initiative of the Deutsche Forschungsgemeinschaft e.V. (DFG), Grant numbers: SCHL 580/3-2, LI 2858/1-2.
- D. Jeannerat and C. Cobas, “Application of multiplet structure deconvolution to extract scalar coupling constants from 1D NMR spectra”, Magn. Reson. Discuss. preprint, in review (2021). https://doi.org/10.5194/mr-2021-32

Tony Davies
Tony Davies is a long-standing Spectroscopy Europe column editor and recognised thought leader on standardisation and regulatory compliance with a foot in both industrial and academic camps. He spent most of his working life in Germany and the Netherlands, most recently as Lead Scientist, Strategic Research Group – Measurement and Analytical Science at AkzoNobel/Nouryon Chemicals BV in the Netherlands. A strong advocate of the correct use of Open Innovation. 0000-0002-3119-4202
0000-0002-3119-4202
[email protected]

Luc Patiny
Luc Patiny has a background in organic chemistry and structural analysis and has been interested in making chemical information available for computers for over 20 years. He is strongly involved in the development of open-source tools, freely accessible and that run directly from a web browser. 0000-0002-4943-2643
0000-0002-4943-2643
[email protected]